Genome Annotation
Genome Annotation
Hu et al. 2019 provided the first ever reference genome of P. nigrum, with 761.2 Mb assembled in 26 pseudochromosomes. The genes assembled in this project were not annotated for protein function; in order to know the function of the different genes of P. nigrum, we performed two different annotation analyses. One analysis was performed using KofamKOALA, which is a KEGG ortholog assignment tool based on profile HMM and adaptive score threshold. With this annotation, a complete metabolome reconstruction was made using KEGG mapper reconstruct tool. The genes were annotated to 3803 different KEGG orthology IDs; 948 of these IDs were part of the metabolic pathways present in the KEGG database (ID: 01100). The complete modules of the metabolic pathways are shown in the next figure.

We found 455 different KEGG orthology IDs for biosynthesis of secondary metabolites pathways (ID: 01110), this can be seen in the next figure.
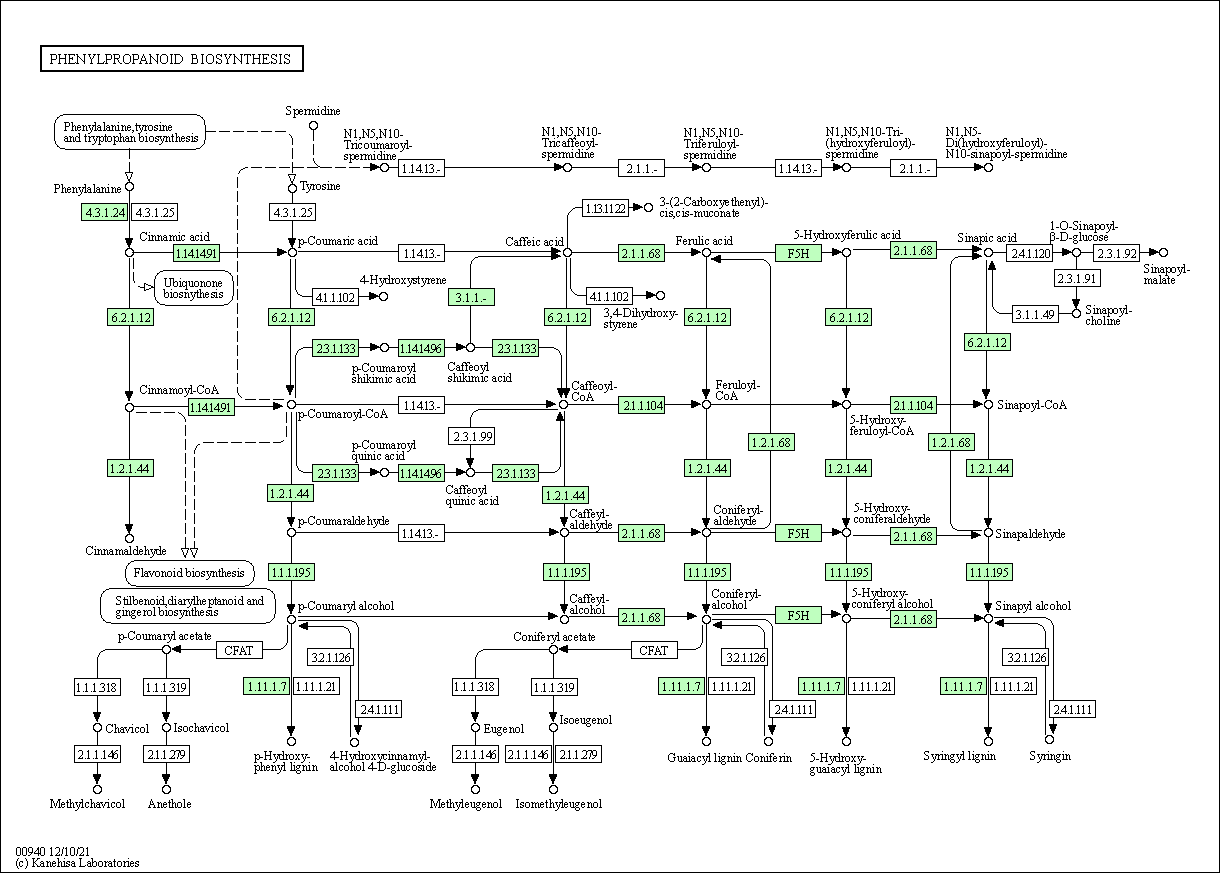
Some of the more interesting pathways for this project are the phenylpropanoid biosynthesis pathway (ID: 00940) and the tropane, piperidine and pyridine alkaloid biosynthesis pathway (ID: 00960). We show the phenylpropanoid biosynthesis pathway, where some enzymes required to obtain ferulic acid from phenylalanine are shown in green; these are KEGG orthology IDs to which some genes from P. nigrum have been annotated.
In the case of the piperidine and pyridine alkaloid biosynthesis pathway, this information is shown in the next figure. In this case, the enzyme of interest is a primary-amine oxidase that performs the reaction that converts cadaverine into 5-Aminopentanal.
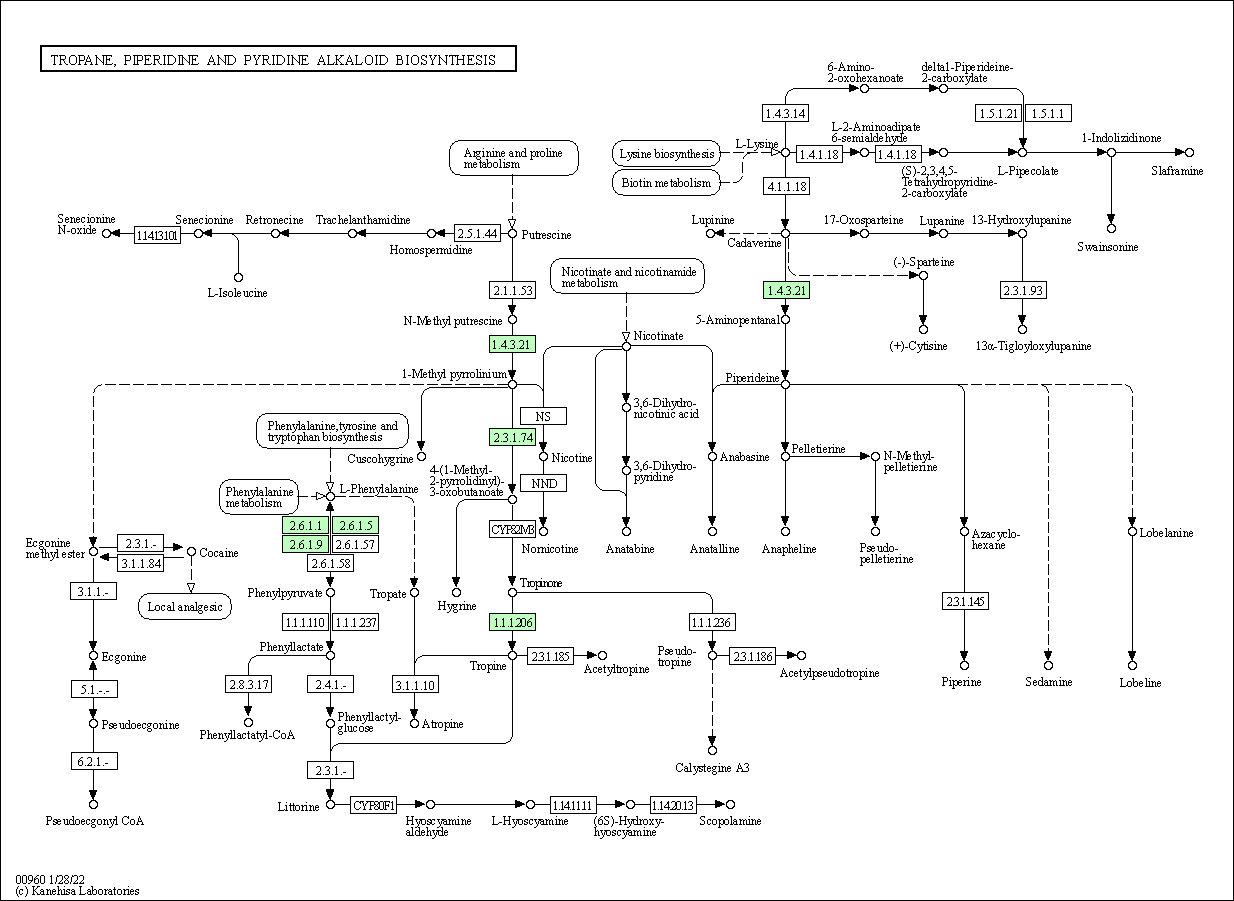
From this annotation, some genes were found that corresponded to the KEGG orthology IDs responsible for performing some steps of the pathway. Pn8.2617 was annotated as a phenylalanine ammonia-lyase, the first enzyme of the proposed pathway; this enzyme was selected as the best candidate for enzyme 1 because it obtained the highest score (1170.9) over the threshold (524.93) for KEGG Orthology ID K10775, with E-value ≊ 0. The second enzyme of the pathway was also found in this way. Pn2.84 was selected as the best candidate for enzyme two of the proposed pathway based on the fact that it obtained the highest score (985.2) over the threshold (555.2) for KEGG Orthology ID K00487, with E-value = 2.5 × 10−297. Pn1.1317 was selected as the best candidate for enzyme number four based on the fact that it obtained the highest score (651.5) over the threshold (499.33) for KEGG Orthology ID K13066, with E-value = 2.9 × 10−196. The eighth enzyme of the reaction, Pn4.3222, was selected based on the fact that it obtained the highest score (889) over the threshold (550.3) for KEGG Orthology ID K00276, with E-value = 2 × 10−267.
The second annotation method used was the GeneFamilyClassifier tool, which is part of PlantTribes, a gene and gene family resource for comparative genomics in plants; this analysis was performed using the Galaxy project platform, a platform for accessible, reproducible and collaborative biomedical analyses. 45,155 genes were annotated for AHRD, TAIR, Pfam domains, InterProScan Descriptions, GO Molecular Functions, Biological Processes and Cellular Components.